jumpincactus
Premium Member
Supporter
- 11,609
- 438
Personal note to farmers....... Use link posted for better function of interactivity of graphs, tables and references.
http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6941.2009.00702.x/full
Keywords:
Abstract
The rhizosphere environment selects a particular microbial community that arises from the one present in bulk soil due to the release of particular compounds in exudates and different opportunities for microbial colonization. During plant–microorganism coevolution, microbial functions supporting plant health and productivity have developed, of which most are described in cultured plant-associated bacteria. This review discusses the state of the art concerning the ecology of the hitherto-uncultured bacteria of the rhizosphere environment, focusing on Acidobacteria, Verrucomicrobia and Planctomycetes. Furthermore, a strategy is proposed to recover bacterial isolates from these taxa from the rhizosphere environment.
Jump to…
Introduction
The rhizosphere, defined here as the volume of soil over which plant roots exert an influence (Hiltner, 1904), differs from bulk soil due to the biophysicochemical processes that occur as a consequence of root growth, water and nutrient uptake, respiration and rhizodeposition (Hinsinger, 1998). The rhizosphere extends from the surface of the plant root to a position in soil that depends on the diffusion rate of exudates and the roots' biochemistry and development (Hinsinger et al., 2005). This extension also varies with the plant type and microbial community composition (Huisman, 1982; Watt et al., 2006b). Besides, as the rhizosphere is known to contain strong gradients of compounds from the root surface into the soil, any rhizosphere sample will inevitably average out the effects of such gradients. In fact, the further away from the root the soil is sampled, the lower the rhizosphere effect on the sample will be. Thus, plant roots create selective pressures that influence the local microbial communities, resulting in effects on their abundance and composition. These selective pressures not only depend on biological processes but also on abiotic ones, such as temperature and water content. For instance, seasonal and daily temperature changes have been found to affect microbial activities (Turpault et al., 2007; Gaumont-Guay et al., 2008) and community composition (Pandey et al., 2001). Water content is also important as it directly influences the microbiota in soils due to the high correlation of microbial activity and soil moisture (Krivtsov et al., 2007). Different studies demonstrated that soil structure, as well as different granulometric fractions determine the presence of different bacterial populations (Sessitsch et al., 2001; Kotani-Tanoi et al., 2007); these factors might influence the microbial communities present in the rhizosphere depending on the soil where the root develops. Also, pH and CO2/O2 tensions will influence the rhizosphere microbial community functional diversity and activity (Jossi et al., 2006; Nelson & Mele, 2006).
The organisms thriving in the rhizosphere encompass a range of different taxa, including prokaryotic and eukaryotic microorganisms. Most abundant among these groups are the bacteria and fungi. Usually, the bacterial fraction ranges from 109 to 1010 cells g−1 of soil and c. 106 cells mm−3 of rhizosphere biofilm in most cases (Watt et al., 2006a). The description of the bacterial taxa present in the rhizosphere will be discussed later.
Key ecological functions can be ascribed to the different bacterial populations that thrive in the rhizosphere environment. For instance, bacteria with nitrogen-fixing and phosphorus-dissolving activities provide nitrogenous compounds and available phosphorus to plant roots, thus increasing the growth of plants, for instance in organic agriculture (Canbolat et al., 2006; Hameeda et al., 2008). Also, the production of particular phytohormones by rhizosphere bacteria can enhance plant growth (Lee & Song, 2007). The activity of the enzyme 1-aminocyclopropane-1-carboxylate deaminase, in particular plant-associated bacteria such as Burkholderia phytofirmans, has important implications for rhizosphere functioning as well (Hardoim et al., 2008). In addition, the rhizosphere microbiota is important, as a wide range of bacteria isolated from this environment have already shown to act as biocontrol agents of plant pathogens, for instance, those producing antimicrobial compounds or eliciting plant defence reactions (Sfalanga et al., 1999; Földes et al., 2000; Jiménez-Esquilín & Roane, 2005; Flores-Vargas & O'Hara, 2006; Romanenko et al., 2007).
The ‘great plate count anomaly’ states that 95–99% of the microbial community present in the environment is not readily accessible by traditional culture techniques (Nichols, 2007). This fraction of the microbial community has been termed as uncultured microbiota. Given the fact that a greater fraction of the total soil microbiota may be culturable than thought before, here we will address this fraction as the hitherto-unculturable bacteria. Most often, the biogeochemical transformations observed in soil have been ascribed to well-characterized culturable bacteria, simply because their ecological relevance has driven a plethora of studies based on culturable bacteria. The large gap between what we know about the culturable microbial diversity vs. the hitherto-unculturable members of these communities is illustrated in Fig. 1. The fraction of culturable members with regard to the total representatives differs for each prokaryotic group. This gap also demonstrates our lack of knowledge of the putative functions hidden in the hitherto-uncultured microbial fractions of particular groups that are involved in rhizospheric microbial–microbial and microbial–plant interactions and pinpoints the need to invest in this basic area of research. One key question is whether members of the uncultured majority are actually involved in key rhizosphere processes. Recently, using stable isotope probing-RNA analysis, a broad new range of inhabitants of the rhizosphere was found that were active under the experimental conditions, but did not have their specific role in the system identified (Vandenkoornhuyse et al., 2007). Moreover, the current ‘omics’ techniques (defined here as integrative studies of biological systems, including genomics, transcriptomics, proteomics and metabolomics) offer a good perspective in describing function or potential function in environmental studies. For instance, proteomics-based studies have already revealed the abundance of particular proteins in the rhizosphere, such as glycoside hydrolases, trypsin/protease inhibitors, plastocyanin-like domains, copper–zinc superoxide dismutases and plant basic secretory proteins (Kiely et al., 2006). Also, metagenomic studies of the rhizospheres of plants growing in acid mine drainage (AMD) revealed several novel microbial genes that are possibly involved in heavy metal resistance to their bacterial hosts (Mirete et al., 2007). Using a metabolomics approach, it was shown that the microbial populations in the rhizosphere were able to metabolize uncommon nutrients exuded by plants (Narasimhan et al., 2003).

Figure 1. Phylogenetic relationship between selected prokaryotic groups represented by 16S rRNA gene sequences of total (culturable and uncultured) bacteria (a) and of culturable-type strains only (b). Selected sequences (1200 bp in length), derived from different ecosystems, originated from the SILVA database, release 94 (Pruesse et al., 2007). Trees were constructed in the arbprogram (Ludwig et al., 2004), and bars indicate a similarity distance of 10% between sequences.
Even though the omics techniques offer great advances in our capabilities to unravel the identity of genes present in the rhizosphere microbiota, there still are technological hurdles that hamper a thorough analysis. For instance, full genomes can only be assembled from dominant species in natural habitats, and only ‘mosaic’, instead of single-species genomes, may be assembled due to the presumed occurrence of closely resembling species in the same habitat (Tyson et al., 2004; Venter et al., 2004). In rhizosphere research, time-course data are of utmost importance due to the drastic changes occurring in associated microbial communities during plant growth. Thus, in cases where shotgun sequencing is used, the sequence information will relate only to a momentary snap shot of the dominant sequences present in the DNA extract, and information on community development in the rhizosphere is often lacking. Also, given the heterogeneity occurring in all natural environments, the interpretation of metagenomics data will be complicated due to the expected variations within residing communities (Tyson et al., 2004; Tringe et al., 2005). Moreover, it is questionable what knowledge of the genome of particular species populations actually means, given the expected genomic heterogeneity within a species, where genomic rearrangements, deletions or insertions may abound (Tringe et al., 2005). Metagenomics will thus provide, at best, information at the level of the prevalence of genes and will depend on the availability of information about the functions of such genes in cultured species (DeLong, 2005; Schirmer et al., 2005; DeLong et al., 2006). The increase in the number of genes with as-yet-unknown functions is keeping pace with the amount of metagenomic information that is becoming available over time. As long as no attempts are made to unravel the function of the many ‘unidentified’ or ‘hypothetical’ proteins commonly found in metagenomic databases, progress in the elucidation of the roles of uncultured microorganisms in ecosystem functioning will be hampered.
In this review, the following questions are posed:
Soils, plant roots and microbial communities – an intricate triplet
Plant–soil–microbial community systems have evolved ever since plants started to colonize the terrestrial environment, around 700 million years ago. The first interactions between soil microbial communities and plant root systems may not have been beneficial to plants, as they may have mainly consisted of attacks to the roots by microorganisms, especially bacteria (Phillips et al., 2003). However, plants colonizing terrestrial environments, being ‘bathed’ in microorganisms, have also initiated evolutionarily useful interactions with microorganisms, for instance, to optimize growth and reproduction. Such – initially primitive – interactions are thought to have resulted in the evolution of all currently known mutually beneficial relationships (Phillips et al., 2003). Classical examples of such interactions are the intricate associations of both rhizobia and mycorrhizal fungi with plant roots. Fossilized remains of primordial root systems also provide evidence that epiphytic and endophytic microbial populations already existed on/in the primitive land plants, although the nature of these associations has not been elucidated (Phillips et al., 2003).
Here we postulate that, in particular highly selected cases, the coevolution of plants and microorganisms in soil has also resulted in firm and dedicated relationships between particular players in the rhizosphere. For one, the belowground competition for soil nutrients between plants certainly has exerted selective pressure that has driven the emergence of plants with wide variations in root structure and function, adapted to perform well under the different conditions in soil. As a consequence, the plant-associated microbial communities may have evolved to the actual scenery of positive, negative and neutral microorganism–microorganism and microorganism–plant interactions. It is known that current-day plant roots and their associated microorganisms are highly affected by soil conditions, which determine the distribution, density and depth of roots in various soils (Passioura, 1991; Jackson et al., 1996; Stewart et al., 1999; Jobbágy & Jackson, 2000).
Thus, firstly, rhizosphere microbial community structure and diversity, in different soil textural types, are strongly determined by the types of plant roots (Garbeva et al., 2004b; Kotani-Tanoi et al., 2007). It is known that during root growth, local soil characteristics, for instance, gas composition and water flow, are increasingly affected by the roots (Colmer, 2003; Lipiec et al., 2007; Benedict & Frelich, 2008; Whalley et al., 2008). In addition, carbonaceous compounds are often abundantly released from roots into the rhizosphere. These factors, in turn, will influence the root-associated microbial communities. Moreover, effects in the opposite way also occur, for example the modulation of root architecture by microorganisms that are locally present, for instance, via the production of phytohormones (Belimov et al., 2009). Some of the relevant interactions between plants and microorganisms, as well as among different microorganisms in the rhizosphere, are summarized in Fig. 2.

Figure 2. Interrelationship between plant, soil and microbial communities. Arrows demonstrate the most relevant interactions occurring between the different players in the rhizosphere.
Many transformations in nutrient cycles that commonly occur in the rhizosphere are still unresolved, whereas others are attributed to microbial activities. Examples of microbial activities in nutrient cycles are, for instance, nitrogen fixation and demineralization (Canbolat et al., 2006) and solubilization of phosphorus (Canbolat et al., 2006) and carbohydrates (Kohler et al., 2006). Plants and other soil microorganisms profit from these transformations occurring in the rhizosphere. Examples of other interactions commonly occurring in the rhizosphere are commensalism, for example by creation of new niches for microorganisms via the secretion of exopolysaccharides (Kaci et al., 2005; Haggag, 2007) or via the production of phytohormones (Lee & Song, 2007), antagonism via production of secondary metabolites with antibiotic activity (Garbeva et al., 2004b; Berg et al., 2005; Costa et al., 2007) and syntrophism, for example via degradation of toxic compounds like recalcitrant hydrocarbons and herbicides (Biryukova et al., 2007; Vaishampayan et al., 2007). Often, a multitude of microbial species is responsible for particular conversions in the rhizosphere and hence a particular degree of functional redundancy may exist for important processes in the rhizosphere. For instance, several different microbial groups are known to be involved in ammonia oxidation in the rhizosphere (Nicolaisen et al., 2004; Herrmann et al., 2008).
However, all of these studies have so far ignored the role of the major part of the hitherto-unculturable soil microbiota. Exploring the hitherto-unculturable bacteria by improvement of culturability presumably will reveal a range of novel functions in the rhizosphere, including functions involved in enhancing rhizosphere competence. An examination of the bacterial groups that are most commonly found in rhizosphere environments and our understanding about their presumed roles in these environments will be discussed further.
Jump to…
Dominant hitherto-uncultured bacteria in the rhizosphere
Soil bacteria with comparable metabolic capacities will respond in a similar fashion to the emergence of different plants, provided that (nutritional) conditions are similar. However, it is still unclear as to what extent commonality exists in the microbial communities that associate with plant roots across plant genera, species or cultivars. This is mostly due to the fact that little attempts were made to synthesize data available in the literature into a systematic evaluation on the relationships between microbial communities, plants, soil types and geographical locations. Given this limitation, we performed such a limited study on data in the literature to indicate the most dominant bacterial groups in the rhizosphere using the following procedure:
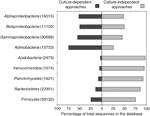
Figure 3. Percentage of 16S rRNA gene sequences from bacterial groups common to the rhizosphere and obtained via culture-dependent and culture-independent approaches. All sequences (1200 bp in length), derived from different ecosystems, were obtained from the Ribosomal Database Project (Coleet al., 2007), release 9.60 (latest accession, 22 April 2008). In parentheses, total number of sequences used per bacterial group.
Within these phyla, the origin of the 16S rRNA gene sequences, i.e. whether these were obtained from isolates or uncultured organisms, was taken into account; it was thus found that the contribution of isolates to the total number of sequences (redundant or not) per bacterial group was variable. For Proteobacteria, the taxonomical information available from isolates was about the same as that from uncultured organisms. However, for Actinobacteria, the available taxonomical information was even higher for isolates, reflecting better adaptation of this group for growth in a pure culture. For the other five groups, by far the highest amount of phylogenetic information was derived from uncultured organisms, i.e. most organisms occurring in these groups are regarded as as-yet-uncultured or plainly uncultured bacteria. Among these groups, the only knowledge with respect to rhizospheric Acidobacteria, Verrucomicrobia and Planctomycetes was obtained on the basis of culture-independent studies. Hence, our understanding of these taxa in the rhizosphere is still limited, as it concerns mainly their 16S rRNA gene fluctuations in the rhizosphere (Sanguin et al., 2006; Zul et al., 2007).
From the three groups, the Acidobacteria and Verrucomicrobia have a low number of species recognized in the literature. To date, the Acidobacteria contain three early-described species, i.e. Acidobacterium capsulatum, Geothrix fermentans and Holophaga foetida (Garrity et al., 2005), next to four recently recognized cultured species (Edaphobacter aggregans, Edaphobacter modestus, Chloracidobacterium thermophilum and Terriglobus roseus (Bryant et al., 2007; Eichorst et al., 2007; Koch et al., 2008). The Verrucomicrobia show a similar picture, with only 10 defined species, i.e. Verrucomicrobium spinosum, four Prosthecobacter spp., Opitutus terrae, Rubritalea marinaand three Xiphinematobacter sp. (Ward-Rainey et al., 1995; Garrity et al., 2005). Although some representatives of the Acidobacteria and Verrucomicrobia have now been cultured from soil (Janssen et al., 2002; Sait et al., 2002; Stevenson et al., 2004; Davis et al., 2005), no isolates have as yet been obtained from the rhizosphere. Isolation directly from the rhizosphere is clearly needed to provide supportive data that these isolates are really rhizosphere-relevant organisms. Their availability would allow an assessment of their influences on plant growth, as well as of their interaction with plants and other soil microorganisms. Also, their involvement in important biogeochemical conversions, degradation of particular compounds and mobilization of nutrients would be facilitated. The members of the Acidobacteria and Verrucomicrobia that have previously been isolated from soil were slow growing, aerobic and heterotrophic bacteria (Janssen et al., 2002; Sait et al., 2002; Stevenson et al., 2004; Davis et al., 2005). Such characteristics are broadly distributed among rhizosphere isolates, which indicates that such isolates may thrive in the rhizosphere environment.
The abundance of the hitherto-unculturable bacteria recognized solely by their phylogeny in the rhizosphere (Kaiser et al., 2001; Kuske et al., 2002; Gremion et al., 2003; Schmalenberger & Tebbe, 2003; Graff & Conrad, 2005; Sharma et al., 2005; Stafford et al., 2005; Sanguin et al., 2006; Wang et al., 2007; Zhang et al., 2007; van Elsas et al., 2008) urges us to address their functional roles. However, information about their putative roles or interactions with the plant is as yet very sparse. Each of the organisms involved might, for instance, occupy a particular niche in the rhizosphere. It is a challenge to develop strategies – based on cultivation-dependent and -independent studies – that allow the exploration of the putative roles and function of the hitherto-unculturable microbiota in the rhizosphere, in particular, analysing their modes of interactions with plants and/or rhizosphere microorganisms.
Jump to…
Ecology of the hitherto-unculturable bacteria in the rhizosphere
If particular niches exist for the hitherto-unculturable bacteria in the rhizosphere, it is logical to assume that selective forces present within such niches act on these rhizosphere populations. Such a selection might be nutritional, dependent on the availability of oxygen or other electron acceptors, or otherwise. To our knowledge, no conclusive data have extensively shown the factors that influence the fate and behaviour of these populations in the rhizosphere. Therefore, the following points should be addressed to elucidate the roles of these organisms in the rhizosphere: (1) their general occurrence and numerical dominance, (2) the ecological conditions driving these populations, (3) in situ (local) metabolic activities and (4) their functional roles in the soil–plant ecosystem.
When comparing bulk and rhizosphere soil, the general occurrence and numerical abundances of the Acidobacteria, Verrucomicrobia and Planctomycetes are often variable, as demonstrated in culture-independent studies based on 16S rRNA gene detection (Dunbar et al., 1999; Kuske et al., 2002; Gremion et al., 2003; Filion et al., 2004; Stafford et al., 2005; Sanguin et al., 2006; De Cárcer et al., 2007; Singh et al., 2007; Zul et al., 2007; Kielak et al., 2008). Specifically, a higher prevalence of 16S rRNA gene clones affiliated with particular groups of Acidobacteria, especially those of groups 1, 2 and 3, was found in rhizospheres of lodgepole pine (Chow et al., 2002),Proteacea sp. (Stafford et al., 2005) and grass (Chow et al., 2002; Stafford et al., 2005; Singh et al., 2007) as compared with corresponding bulk soils. Also, a higher number of 16S rRNA gene clones of Verrucomicrobia was observed in rhizospheres of lodgepole pine, particularly subdivisions 2, 3 and 4 (Chow et al., 2002), Thlaspi caerulescens, subdivision 2 (Gremion et al., 2003) and Proteacea sp., subdivision 3 (Stafford et al., 2005) in comparison with the corresponding bulk soils. Besides, Planctomycetesrepresentatives were found in higher numbers in the rhizospheres of T. caerulescens than in the bulk soil, with 16S rRNA gene clones affiliating only with the Nostocoida limilula III cluster (Gremion et al., 2003) and Proteacea sp. (Stafford et al., 2005) (Table 1). In contrast, a study performed with different plants demonstrated higher numbers of Acidobacteria in a 16S rRNA gene clone library made from bulk soil DNA than in the ones made from rhizosphere DNA, although no information was given about the dominant groups in both soil compartments (Kielak et al., 2008). Also, the rhizosphere of maize plants showed lower numbers of Acidobacteria, Verrucomicrobia andPlanctomycetes than the corresponding bulk soil, and again no information was given about the dominant subdivisions (Sanguin et al., 2006). In T. caerulescens (Gremionet al., 2003), different Acidobacteria groups were found in bulk and rhizosphere soil, although the number of detected 16S rRNA genes was the same in both soil compartments. In this study, more clones were found to be affiliated with Acidobacteria subdivision 6 in the rhizosphere, whereas in the bulk soil more clones were found to be affiliated with Acidobacteria subdivision 1. Furthermore, the diversities of the Acidobacteria and Verrucomicrobia in the rhizosphere of Lolium perenne L. were large. Such diversities even approximated those of the total Proteobacteria (Singh et al., 2007).
Table 1. Abundance of Acidobacteria, Verrucomicrobia and Planctomycetes and presumed factors influencing community sizes in the rhizosphere
Plant species Soil type Approach Major observations made References
Higher abundance of the hitherto- uncultured groups found at Presumed factors influencing the Acidobacteria,Verrucomicrobia andPlanctomycetes communities
Bulk soil Rhizosphere
Bunchgrasses (Stipa hymenoides andHilaria jamesii); grass (Bromus tectorum) Calcareous loamy sand B ND ND • The composition ofAcidobacterium division members differed with soil depth and with the presence of plants or cyanobacterial crust on the top soil layer Kuske et al.(2002)
Thlaspi caerulescens Sandy loam A VerrucomicrobiaandPlanctomycetes • Plant roots exerted a strong selective pressure on the diversity of Acidobacteria, Verrucomicrobiaand Planctomycetesrepresentatives Gremion et al. (2003)
Conifer (Picea mariana) Not described C ND ND • Representatives of the division ofAcidobacteria were higher in number and diversity in the rhizosphere of healthy plants than of plants with black spruce •Verrucomicrobia representatives were only found in the rhizosphere of diseased black spruce plants Filion et al.(2004)
Proteacea species (Leucospermun truncatulum andLecadendron xanthoconus) Acid sand-stone-derived soil A Acidobacteria, VerrucomicrobiaandPlanctomycetes • Endemic communities in the rhizosphere differed per geographic location and soil covering vegetation type Stafford et al. (2005)
Maize (Zea mays cv. PR38a24) Sand–loam soil D Acidobacteria, VerrucomicrobiaandPlanctomycetes • Plant roots exerted a strong selective pressure on the diversity of Acidobacteria, Verrucomicrobiaand Planctomycetesrepresentatives Sanguin et al. (2006)
Willow (Salix vimanalis×schewerinii) Not described E ND ND • PCB had no effect on theAcidobacteria diversity in the rhizosphere of Willow De Cárcer et al. (2007)
Grass (Lolium perenne L.) Sand–silt soil A Acidobacteria • Acidobacterial communities were favoured in rhizosphere soil, but the major determinant of community composition was soil type Singh et al.(2007)
Different plant communities Endostagnic cambisol soil F Verrucomicrobia, PlanctomycetesandAcidobacteria • Plant species strongly influenced the bacterial community composition; season was identified as a second important factor. No effects caused by plant diversity or water content were observed Zul et al.(2007)
Different plant diversity treatments Loamy sand soil A Acidobacteria Verrucomicrobia • No detectable changes in acidobacterial or verrucomicrobial population diversity and structure were observed in response to different vegetation coverages Kielak et al.(2008)
Yew (Taxus×mediaand Taxus mairei) Not described A ND ND • Geographic origin is more important in shaping the bacterial community composition than plant species Hao et al.(2008)
Wild oat root (Avena fatua) Medium-texture loam G Acidobacteria, VerrucomicrobiaandPlanctomycetes • Rhizosphere effect increased numbers of Acidobacteria, Verrucomicrobia andPlanctomycetes member in rhizosphere soil when compared with the respective bulk soil • The number of Acidobacteria, Verrucomicrobia andPlancomycetes also differed among root tip, root hairs and mature root DeAngelis et al. (2009)
The numerical abundance of members of the Acidobacteria, Verrucomicrobia and Planctomycetes is not only dependent on differences between bulk and rhizosphere soil but also changes are incited upon root development. In a microcosm experiment performed with wild oat roots, members of the Acidobacteria, Verrucomicrobia andPlanctomycetes occurred at a higher number, as demonstrated by the higher number of 16S rRNA gene copies in the rhizosphere of adult plants than in the bulk soil (DeAngelis et al., 2009). Besides, there was a greater abundance of Acidobacteria 16S rRNA genes at the surface of mature roots than at root hairs and tips; thePlanctomycetes showed higher 16S rRNA gene abundance at the surface of mature roots and root tips, than at the root hairs, and the Verrucomicrobia, although having a lower abundance at the root tips, had greater abundance at the surface of mature roots and root hairs than in the corresponding bulk soil (DeAngelis et al., 2009).
Clearly, the high diversity of acidobacterial and verrucomicrobial species indicates that different bacterial populations are favoured by different selective forces exerted in the rhizosphere. At the microhabitat level, such forces can be different across space and time, thus allowing the emergence of diverse populations across each phylum. An important question here is which selective forces influence the acidobacterial, verrucomicrobial and Planctomycetales communities in the rhizosphere. On the basis of incidental data, this question has been difficult to answer. Thus, no consensus was found about which ecological factors would select for these organisms in the rhizosphere. Recent studies demonstrated that different factors select distinct members of these groups (Table 1). For instance, plant species (Chow et al., 2002; Gremionet al., 2003; Sanguin et al., 2006), soil type (Singh et al., 2007), field history (Kielak et al., 2008) and different abiotic factors (Stafford et al., 2005; Hao et al., 2008) have all been implicated as selective factors that differentially affect these groups (Table 1).
Moreover, another key question is whether we can actually show to what extent hitherto-uncultured organisms, for example members of the Acidobacteria, are part of the metabolically active community (derived from the higher abundances in RNA vs. DNA extracts) in the rhizosphere and how this is temporally organized. In the metal-hyperaccumulating plant T. caerulescens, a major discrepancy in abundance was found between the metabolically active and the total Acidobacteria (Gremion et al., 2003). In this study, it was indicated that only a subset of the Acidobacteria might be active in the rhizosphere, whereas the majority may be inactive or dormant. On the other hand, in a study using a culture-independent approach to search for 16S rRNA gene sequences in DNA and RNA extracts from the chestnut tree (Castanea crenata) rhizosphere (Lee et al., 2008), Acidobacteria were found to be not only numerically dominant but also metabolically active, implying that these Acidobacteria may play a key role in this habitat. Additionally, in a metagenomic study conducted on the rhizosphere of Erica andevalensis adapted to AMD, it was demonstrated that in this acid environment,Acidobacteria of subdivision 1 were metabolically active (Mirete et al., 2007). The authors also found that four of 13 clones with nickel resistance genes most likely were from representatives of Acidobacteria subdivision 1 (Mirete et al., 2007). However, it is unclear whether these genes were actually expressed in the environment and whether they play key roles in the heavy metal resistance of E. andevalensis.
Jump to…
Methods to improve bacterial and archaeal culturability
Recently, several successes in the culturing of the hitherto-uncultured bacteria were reported (Table 2). Key factors that presumably influence bacterial colony formation on solid media were found to be the type of growth medium, the incubation conditions (temperature, CO2 concentration and time of incubation) and several factors that are known to reduce oxidative stress (e.g. the presence of catalase or pyruvate) (Stevenson et al., 2004; Van Overbeek et al., 2004; Sangwan et al., 2005; Eichorst et al., 2007). It is a well-known fact that medium composition will determine which fraction of the microbial community is recoverable from the environment. Often, the medium itself limits bacterial growth due to the presence of excessive amounts of nutrients that do not reflect environmental conditions (Janssen et al., 2002; Sait et al., 2002). This phenomenon, known as ‘substrate-accelerated death’, has first been observed when organisms growing under oligotrophic conditions are transferred to high substrate concentrations (Postgate & Hunter, 1964; Straskrabová, 1983). It is a key issue, given the roughly low substrate concentrations in the rhizosphere compartment. Therefore, to overcome this limitation, novel media are designed to match conditions in the environment in which some of the key facets (e.g. amount of nutrients, nutrient composition, the presence of trace elements and pH) are adjusted for optimized bacterial growth. For instance, the use of soil-extract agar medium (Pochon & Tardieu, 1962; Hamaki et al., 2005) or of dilute standard media, both aiming to reduce nutrient levels in order to avoid substrate-accelerated death, has allowed the isolation of bacterial groups that had been regarded as uncultured only a short while ago (Joseph et al., 2003; Davis et al., 2005). Also, the presence of fast-growing bacteria that inhibit colony formation of slower growing ones, especially on nutrient-rich media, affects the recovery of hard-to-culture organisms. Media with low nutrient levels, in combination with long incubation periods at relatively low temperatures, will allow colony formation by bacteria that have low growth rates. The approach has been successful in revealing a range of novel organisms in various recent studies (Janssen et al., 2002; Rappe et al., 2002; Sait et al., 2002; Davis et al., 2005; Miteva & Brenchley, 2005; Sangwan et al., 2005; Eichorst et al., 2007). Hence, this allowed the cultivation from soil of a range of novel organisms such as bacteria belonging to the following groups: Chloroflexi, Planctomycetes, Acidobacteria, Verrucomicrobia and some Archaea representatives (Table 2). However, such approaches have seldom been used with rhizosphere samples, and only one reference was found reporting cultivation of hard-to-culture microorganisms from rhizosphere samples (Simon et al., 2005).
Table 2. Factors hampering bacterial growth upon isolation from natural environments and proposed approaches to improve culturability
Factors that limit growth of the hitherto-uncultured bacteria Proposed approaches to improve culturability Phylogeny of novel isolates that were obtained Environments from where novel isolates were obtained References
Inability to grow at high nutrient concentrations and overgrowth by faster growing species • Reduce nutrient availability in growth medium • Apply a longer incubation time Actinobacteria, Acidobacteria, Proteobacteria, Verrucomicrobia, Gammaproteobacteria, SAR11 clade,Gemmatimonadetes, Chloroflexi, Planctomycetes, CFB group • Bulk soil Janssen et al. (2002);Sait et al. (2002);Davis et al. (2005);Sangwan et al. (2005);Eichorst et al. (2007)
• Seawater Rappe et al. (2002);Miteva & Brenchley (2005)
Media selectiveness for particular groups of microorganisms • Development of media that select for a different or a broader spectrum of microorganisms Acidobacteria, Gemmatimonadetes, Chloroflexi, Alphaproteobacteria, Betaproteobacteria, Planctomycetes, Gammaproteobacteria, Actinobacteria, • Bulk soil Joseph et al. (2003);Davis et al. (2005);Hamaki et al. (2005)
Compounds inhibiting bacterial growth • Diffusion or dilution of growth-inhibiting compounds, for example, by making use of diffusion chambers Deltaproteobacteria, Verrucomicrobia, Spirochaetes, Acidobacteria, Planctomycetes, Proteobacteria, Bacteroidetes • Marine sediment Bollmann et al. (2007)
• Fresh water sediment Tamaki et al. (2005)
• Application of alternative solidifying agents, for example gellan gun instead of agar
Syntrophic growth or requirement for growth factors produced by other microorganisms • Incubation of environmental samples in diffusion chambers • Addition of syntrophic bacteria in enrichment cultures Deltaproteobacteria, Verrucomicrobia, Spirochaetes, Acidobacteria Rice Cluster-I methanogen group Archaea • Rice paddy field Sakai et al. (2007)
• Fresh water sediment Bollmann et al. (2007)
• Bulk soil Cavaletti et al. (2006)
• Rhizosphere Simon et al. (2005)
CO2 tension during incubation • Increase CO2 tension during incubation Verrucomicrobia, Acidobacteria • Bulk soil Stevenson et al.(2004); Sangwan et al.(2005); Eichorst et al.(2007)
Low abundance in environmental samples • Single-cell detection combined with parallel microbial cultivation • High-throughput dilution to extinction cultivation method • Micro-Petri dish • Bulk soil Zengler et al. (2005)
• Seawater Rappe et al. (2002)
• Fresh water Ingham et al. (2007)
Formation of colonies that are undetectable by the unarmed eye • Colony identification by FISH in microtitre plates • Colony identification by hybridization on nylon membranes • Microcolony cultivation Verrucomicrobia, Candidate division TM7, Acidobacteria • Bulk soil Stevenson et al.(2004); Ferrari et al.(2005); Sangwan et al.(2005)
• Sea water Rappe et al. (2002)
Restriction of bacterial growth upon the presence of reactive oxygen species • Reduction of oxidative stress by addition of oxidation-protective agents Acidobacteria • Bulk soil Stevenson et al.(2004)
Lack of ability to grow under specific circumstances, i.e. growth on solid or in a liquid substrate • Screening for isolates in both liquid and solid media Acidobacteria, Verrucomicrobia, Gemmatimonadetes, Proteobacteriaand CFB group • Sea food, salt-processed food, raw vegetables Shigematsu et al.(2007)
• Seawater Miteva & Brenchley (2005); Simu et al.(2005)
• Bulk soil Schoenborn et al.(2004)
Besides, the inadvertent presence of growth-inhibitory compounds in agar media may prevent the formation of colonies by particular bacteria. The use of gellan gum as an alternative solidifying agent was shown to allow the cultivation of many hitherto-unculturable bacteria (Tamaki et al., 2005), as the agar might actually be inhibitory to some groups of microorganisms, for example, those from freshwater. Also, incubation of samples (in this case from marine sediments) in diffusion chambers, which allows dilution of inhibitory compounds produced by bacteria during growth, permitted the growth of novel bacterial groups (Bollmann et al., 2007) (Table 2). Another reason why many bacteria cannot be cultured is the obligate growth of bacteria in consortia (Sakai et al., 2007). Thus, the growth of individual species in these consortia relies on the growth of others, for example when bacteria feed on compounds produced by others (Simon et al., 2005) or require growth factors produced by other bacteria (Bollmann et al., 2007). Therefore, it will be relevant to screen for unknown species in clone libraries made from bacterial mixtures, for example, by plating undiluted rhizosphere samples.
Jump to…
Towards the as-yet-uncultured microbial populations in the rhizosphere and their basic ecology
The recovery of the broadest spectrum of species diversity from the rhizosphere will offer a new insight into hitherto unculturable populations and their functions, as demonstrated with the recovery of novel nickel resistance genes from heather (E. andevalensis) rhizosphere metagenome (Mirete et al., 2007). A major focus should be placed on those bacterial phyla that are presumably abundant in the rhizosphere and that have so far only been described via culture-independent approaches, i.e. theAcidobacteria, Verrucomicrobia and Planctomycetes. Several studies have indicated strategies to improve culturability in natural ecosystems (Table 2), but only one has so far been addressed in the rhizosphere, in this case by improving the culturability of Archaea species (Simon et al., 2005).
Improving the cultivation of Acidobacteria and Verrucomicrobia from the rhizosphere
The rhizosphere clearly is a selective habitat for different groups of microorganisms in soil and this selectivity comes about as a result of a plethora of beneficial and stress conditions imposed on its inhabitants. Analysis of the factors that shape rhizosphere conditions might help to design methods for the recovery of a broad range of novel isolates. The following factors should be considered as relevant for the design of such methods: (1) the provision of the (solid) substrate for bacterial growth; (2) the nutritional status of the rhizosphere, which is often oligotrophic, although pulse-wise receiving nutrients; (3) the high partial CO2 and low partial O2 pressures (due to bacterial and root respiration); (4) the temporary and fluctuating lowering of the pH and (5) the presence of signalling and other (eventually toxic) molecules of root and bacterial origin. Given the fact that no single medium or technique is likely to solve the culturability issue all at once, here, a polyphasic approach is proposed, which should apply parallel cultivation systems in an attempt to yield as many hitherto-uncultured bacteria from the rhizosphere as possible. Key to such a polyphasic approach will be:
Jump to…
Conclusions
Rhizosphere communities are important for key plant processes such as nutrient acquisition, protection against soil-borne diseases and plant development, as the rhizosphere is the transitional zone in between bulk soil and plant roots. The key functions of rhizosphere bacterial communities, for instance nitrogen fixation (Canbolat et al., 2006), phosphate solubilization (Kohler et al., 2006) and plant health protection (Garbeva et al., 2004a), have been described, mainly for well-characterized culturable species.
In this review, we show that among the groups of bacteria highest in density in the rhizosphere environment, the Acidobacteria, Verrucomicrobia and Planctomycetes are the ones whose functions remain to be unveiled. So far, these three groups have not been cultivated from this environment. Therefore, most of the information that emerged on these taxa in the rhizosphere was from measurements on the 16S rRNA gene level. The only exception is a metagenomics study conducted in the rhizosphere of plants adapted to AMD (Mirete et al., 2007). However, in this study, not all modes of action of the novel genes found could be elucidated nor was it shown that these genes were actually active in the rhizosphere environment.
One way to circumvent the hurdles of culture-independent techniques (Van Elsas et al., 2008) is the development of approaches that improve culturability, here, with a special focus on the rhizosphere environment. Several studies already aimed at the isolation of novel species from bulk soil, with variable successes in the recovery ofAcidobacteria and Verrucomicrobia. Although these two groups have shown their recalcitrance to cultivation from the rhizosphere environment, it is likely to presume that they can be isolated using approaches directed to recover the widest possible range of bacteria. We propose the use of a polyphasic approach comprising low-nutrient solid and liquid media with the inclusion of oxidative stress-protective agents and/or rhizosphere extract, and long incubation periods at elevated carbon dioxide concentrations. The exploration of hitherto-uncultured organisms – following their growth to purity – will clearly simplify an assessment of their interactions with other organisms, which cannot be achieved via culture-independent approaches.
Moreover, there is a lack of studies that merge culture-dependent with advanced culture-independent studies. Such a combination of approaches will greatly increase our analytical power in order to answer one of the ultimate questions in microbial ecology –‘Which are the ecological niches, putative roles, functions and modes of interactions of the hitherto-uncultured microbiota?’
http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6941.2009.00702.x/full
References
http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6941.2009.00702.x/full
Keywords:
- rhizosphere;
- hitherto-uncultured bacteria;
- Acidobacteria;
- Verrucomicrobia;
- improving culturability
Abstract
The rhizosphere environment selects a particular microbial community that arises from the one present in bulk soil due to the release of particular compounds in exudates and different opportunities for microbial colonization. During plant–microorganism coevolution, microbial functions supporting plant health and productivity have developed, of which most are described in cultured plant-associated bacteria. This review discusses the state of the art concerning the ecology of the hitherto-uncultured bacteria of the rhizosphere environment, focusing on Acidobacteria, Verrucomicrobia and Planctomycetes. Furthermore, a strategy is proposed to recover bacterial isolates from these taxa from the rhizosphere environment.
Jump to…
Introduction
The rhizosphere, defined here as the volume of soil over which plant roots exert an influence (Hiltner, 1904), differs from bulk soil due to the biophysicochemical processes that occur as a consequence of root growth, water and nutrient uptake, respiration and rhizodeposition (Hinsinger, 1998). The rhizosphere extends from the surface of the plant root to a position in soil that depends on the diffusion rate of exudates and the roots' biochemistry and development (Hinsinger et al., 2005). This extension also varies with the plant type and microbial community composition (Huisman, 1982; Watt et al., 2006b). Besides, as the rhizosphere is known to contain strong gradients of compounds from the root surface into the soil, any rhizosphere sample will inevitably average out the effects of such gradients. In fact, the further away from the root the soil is sampled, the lower the rhizosphere effect on the sample will be. Thus, plant roots create selective pressures that influence the local microbial communities, resulting in effects on their abundance and composition. These selective pressures not only depend on biological processes but also on abiotic ones, such as temperature and water content. For instance, seasonal and daily temperature changes have been found to affect microbial activities (Turpault et al., 2007; Gaumont-Guay et al., 2008) and community composition (Pandey et al., 2001). Water content is also important as it directly influences the microbiota in soils due to the high correlation of microbial activity and soil moisture (Krivtsov et al., 2007). Different studies demonstrated that soil structure, as well as different granulometric fractions determine the presence of different bacterial populations (Sessitsch et al., 2001; Kotani-Tanoi et al., 2007); these factors might influence the microbial communities present in the rhizosphere depending on the soil where the root develops. Also, pH and CO2/O2 tensions will influence the rhizosphere microbial community functional diversity and activity (Jossi et al., 2006; Nelson & Mele, 2006).
The organisms thriving in the rhizosphere encompass a range of different taxa, including prokaryotic and eukaryotic microorganisms. Most abundant among these groups are the bacteria and fungi. Usually, the bacterial fraction ranges from 109 to 1010 cells g−1 of soil and c. 106 cells mm−3 of rhizosphere biofilm in most cases (Watt et al., 2006a). The description of the bacterial taxa present in the rhizosphere will be discussed later.
Key ecological functions can be ascribed to the different bacterial populations that thrive in the rhizosphere environment. For instance, bacteria with nitrogen-fixing and phosphorus-dissolving activities provide nitrogenous compounds and available phosphorus to plant roots, thus increasing the growth of plants, for instance in organic agriculture (Canbolat et al., 2006; Hameeda et al., 2008). Also, the production of particular phytohormones by rhizosphere bacteria can enhance plant growth (Lee & Song, 2007). The activity of the enzyme 1-aminocyclopropane-1-carboxylate deaminase, in particular plant-associated bacteria such as Burkholderia phytofirmans, has important implications for rhizosphere functioning as well (Hardoim et al., 2008). In addition, the rhizosphere microbiota is important, as a wide range of bacteria isolated from this environment have already shown to act as biocontrol agents of plant pathogens, for instance, those producing antimicrobial compounds or eliciting plant defence reactions (Sfalanga et al., 1999; Földes et al., 2000; Jiménez-Esquilín & Roane, 2005; Flores-Vargas & O'Hara, 2006; Romanenko et al., 2007).
The ‘great plate count anomaly’ states that 95–99% of the microbial community present in the environment is not readily accessible by traditional culture techniques (Nichols, 2007). This fraction of the microbial community has been termed as uncultured microbiota. Given the fact that a greater fraction of the total soil microbiota may be culturable than thought before, here we will address this fraction as the hitherto-unculturable bacteria. Most often, the biogeochemical transformations observed in soil have been ascribed to well-characterized culturable bacteria, simply because their ecological relevance has driven a plethora of studies based on culturable bacteria. The large gap between what we know about the culturable microbial diversity vs. the hitherto-unculturable members of these communities is illustrated in Fig. 1. The fraction of culturable members with regard to the total representatives differs for each prokaryotic group. This gap also demonstrates our lack of knowledge of the putative functions hidden in the hitherto-uncultured microbial fractions of particular groups that are involved in rhizospheric microbial–microbial and microbial–plant interactions and pinpoints the need to invest in this basic area of research. One key question is whether members of the uncultured majority are actually involved in key rhizosphere processes. Recently, using stable isotope probing-RNA analysis, a broad new range of inhabitants of the rhizosphere was found that were active under the experimental conditions, but did not have their specific role in the system identified (Vandenkoornhuyse et al., 2007). Moreover, the current ‘omics’ techniques (defined here as integrative studies of biological systems, including genomics, transcriptomics, proteomics and metabolomics) offer a good perspective in describing function or potential function in environmental studies. For instance, proteomics-based studies have already revealed the abundance of particular proteins in the rhizosphere, such as glycoside hydrolases, trypsin/protease inhibitors, plastocyanin-like domains, copper–zinc superoxide dismutases and plant basic secretory proteins (Kiely et al., 2006). Also, metagenomic studies of the rhizospheres of plants growing in acid mine drainage (AMD) revealed several novel microbial genes that are possibly involved in heavy metal resistance to their bacterial hosts (Mirete et al., 2007). Using a metabolomics approach, it was shown that the microbial populations in the rhizosphere were able to metabolize uncommon nutrients exuded by plants (Narasimhan et al., 2003).

Figure 1. Phylogenetic relationship between selected prokaryotic groups represented by 16S rRNA gene sequences of total (culturable and uncultured) bacteria (a) and of culturable-type strains only (b). Selected sequences (1200 bp in length), derived from different ecosystems, originated from the SILVA database, release 94 (Pruesse et al., 2007). Trees were constructed in the arbprogram (Ludwig et al., 2004), and bars indicate a similarity distance of 10% between sequences.
Even though the omics techniques offer great advances in our capabilities to unravel the identity of genes present in the rhizosphere microbiota, there still are technological hurdles that hamper a thorough analysis. For instance, full genomes can only be assembled from dominant species in natural habitats, and only ‘mosaic’, instead of single-species genomes, may be assembled due to the presumed occurrence of closely resembling species in the same habitat (Tyson et al., 2004; Venter et al., 2004). In rhizosphere research, time-course data are of utmost importance due to the drastic changes occurring in associated microbial communities during plant growth. Thus, in cases where shotgun sequencing is used, the sequence information will relate only to a momentary snap shot of the dominant sequences present in the DNA extract, and information on community development in the rhizosphere is often lacking. Also, given the heterogeneity occurring in all natural environments, the interpretation of metagenomics data will be complicated due to the expected variations within residing communities (Tyson et al., 2004; Tringe et al., 2005). Moreover, it is questionable what knowledge of the genome of particular species populations actually means, given the expected genomic heterogeneity within a species, where genomic rearrangements, deletions or insertions may abound (Tringe et al., 2005). Metagenomics will thus provide, at best, information at the level of the prevalence of genes and will depend on the availability of information about the functions of such genes in cultured species (DeLong, 2005; Schirmer et al., 2005; DeLong et al., 2006). The increase in the number of genes with as-yet-unknown functions is keeping pace with the amount of metagenomic information that is becoming available over time. As long as no attempts are made to unravel the function of the many ‘unidentified’ or ‘hypothetical’ proteins commonly found in metagenomic databases, progress in the elucidation of the roles of uncultured microorganisms in ecosystem functioning will be hampered.
In this review, the following questions are posed:
- 1
What are the key interactions among soil, plant and the local microbial communities? - 2
Which are the dominant hitherto-unculturable bacteria in the rhizosphere environment? - 3
Can rhizosphere-relevant functions of these organisms in their natural habitat be revealed by the actual knowledge in the literature? - 4
How to culture the hitherto-unculturable bacteria from rhizosphere environment? - 5
How to reveal the basic ecology of the hitherto-unculturable populations in the rhizosphere?
Soils, plant roots and microbial communities – an intricate triplet
Plant–soil–microbial community systems have evolved ever since plants started to colonize the terrestrial environment, around 700 million years ago. The first interactions between soil microbial communities and plant root systems may not have been beneficial to plants, as they may have mainly consisted of attacks to the roots by microorganisms, especially bacteria (Phillips et al., 2003). However, plants colonizing terrestrial environments, being ‘bathed’ in microorganisms, have also initiated evolutionarily useful interactions with microorganisms, for instance, to optimize growth and reproduction. Such – initially primitive – interactions are thought to have resulted in the evolution of all currently known mutually beneficial relationships (Phillips et al., 2003). Classical examples of such interactions are the intricate associations of both rhizobia and mycorrhizal fungi with plant roots. Fossilized remains of primordial root systems also provide evidence that epiphytic and endophytic microbial populations already existed on/in the primitive land plants, although the nature of these associations has not been elucidated (Phillips et al., 2003).
Here we postulate that, in particular highly selected cases, the coevolution of plants and microorganisms in soil has also resulted in firm and dedicated relationships between particular players in the rhizosphere. For one, the belowground competition for soil nutrients between plants certainly has exerted selective pressure that has driven the emergence of plants with wide variations in root structure and function, adapted to perform well under the different conditions in soil. As a consequence, the plant-associated microbial communities may have evolved to the actual scenery of positive, negative and neutral microorganism–microorganism and microorganism–plant interactions. It is known that current-day plant roots and their associated microorganisms are highly affected by soil conditions, which determine the distribution, density and depth of roots in various soils (Passioura, 1991; Jackson et al., 1996; Stewart et al., 1999; Jobbágy & Jackson, 2000).
Thus, firstly, rhizosphere microbial community structure and diversity, in different soil textural types, are strongly determined by the types of plant roots (Garbeva et al., 2004b; Kotani-Tanoi et al., 2007). It is known that during root growth, local soil characteristics, for instance, gas composition and water flow, are increasingly affected by the roots (Colmer, 2003; Lipiec et al., 2007; Benedict & Frelich, 2008; Whalley et al., 2008). In addition, carbonaceous compounds are often abundantly released from roots into the rhizosphere. These factors, in turn, will influence the root-associated microbial communities. Moreover, effects in the opposite way also occur, for example the modulation of root architecture by microorganisms that are locally present, for instance, via the production of phytohormones (Belimov et al., 2009). Some of the relevant interactions between plants and microorganisms, as well as among different microorganisms in the rhizosphere, are summarized in Fig. 2.

Figure 2. Interrelationship between plant, soil and microbial communities. Arrows demonstrate the most relevant interactions occurring between the different players in the rhizosphere.
Many transformations in nutrient cycles that commonly occur in the rhizosphere are still unresolved, whereas others are attributed to microbial activities. Examples of microbial activities in nutrient cycles are, for instance, nitrogen fixation and demineralization (Canbolat et al., 2006) and solubilization of phosphorus (Canbolat et al., 2006) and carbohydrates (Kohler et al., 2006). Plants and other soil microorganisms profit from these transformations occurring in the rhizosphere. Examples of other interactions commonly occurring in the rhizosphere are commensalism, for example by creation of new niches for microorganisms via the secretion of exopolysaccharides (Kaci et al., 2005; Haggag, 2007) or via the production of phytohormones (Lee & Song, 2007), antagonism via production of secondary metabolites with antibiotic activity (Garbeva et al., 2004b; Berg et al., 2005; Costa et al., 2007) and syntrophism, for example via degradation of toxic compounds like recalcitrant hydrocarbons and herbicides (Biryukova et al., 2007; Vaishampayan et al., 2007). Often, a multitude of microbial species is responsible for particular conversions in the rhizosphere and hence a particular degree of functional redundancy may exist for important processes in the rhizosphere. For instance, several different microbial groups are known to be involved in ammonia oxidation in the rhizosphere (Nicolaisen et al., 2004; Herrmann et al., 2008).
However, all of these studies have so far ignored the role of the major part of the hitherto-unculturable soil microbiota. Exploring the hitherto-unculturable bacteria by improvement of culturability presumably will reveal a range of novel functions in the rhizosphere, including functions involved in enhancing rhizosphere competence. An examination of the bacterial groups that are most commonly found in rhizosphere environments and our understanding about their presumed roles in these environments will be discussed further.
Jump to…
Dominant hitherto-uncultured bacteria in the rhizosphere
Soil bacteria with comparable metabolic capacities will respond in a similar fashion to the emergence of different plants, provided that (nutritional) conditions are similar. However, it is still unclear as to what extent commonality exists in the microbial communities that associate with plant roots across plant genera, species or cultivars. This is mostly due to the fact that little attempts were made to synthesize data available in the literature into a systematic evaluation on the relationships between microbial communities, plants, soil types and geographical locations. Given this limitation, we performed such a limited study on data in the literature to indicate the most dominant bacterial groups in the rhizosphere using the following procedure:
- 1
Collation of an inventory of culture-independent studies that addresses the rhizosphere bacterial diversity across a broad array of plant types. - 2
Identification of the most dominant phyla in the rhizosphere as found in each study. - 3
Singling out the most dominant groups as the ‘most commonly found’ taxa in the rhizosphere by frequency of detection across all studies.

Figure 3. Percentage of 16S rRNA gene sequences from bacterial groups common to the rhizosphere and obtained via culture-dependent and culture-independent approaches. All sequences (1200 bp in length), derived from different ecosystems, were obtained from the Ribosomal Database Project (Coleet al., 2007), release 9.60 (latest accession, 22 April 2008). In parentheses, total number of sequences used per bacterial group.
Within these phyla, the origin of the 16S rRNA gene sequences, i.e. whether these were obtained from isolates or uncultured organisms, was taken into account; it was thus found that the contribution of isolates to the total number of sequences (redundant or not) per bacterial group was variable. For Proteobacteria, the taxonomical information available from isolates was about the same as that from uncultured organisms. However, for Actinobacteria, the available taxonomical information was even higher for isolates, reflecting better adaptation of this group for growth in a pure culture. For the other five groups, by far the highest amount of phylogenetic information was derived from uncultured organisms, i.e. most organisms occurring in these groups are regarded as as-yet-uncultured or plainly uncultured bacteria. Among these groups, the only knowledge with respect to rhizospheric Acidobacteria, Verrucomicrobia and Planctomycetes was obtained on the basis of culture-independent studies. Hence, our understanding of these taxa in the rhizosphere is still limited, as it concerns mainly their 16S rRNA gene fluctuations in the rhizosphere (Sanguin et al., 2006; Zul et al., 2007).
From the three groups, the Acidobacteria and Verrucomicrobia have a low number of species recognized in the literature. To date, the Acidobacteria contain three early-described species, i.e. Acidobacterium capsulatum, Geothrix fermentans and Holophaga foetida (Garrity et al., 2005), next to four recently recognized cultured species (Edaphobacter aggregans, Edaphobacter modestus, Chloracidobacterium thermophilum and Terriglobus roseus (Bryant et al., 2007; Eichorst et al., 2007; Koch et al., 2008). The Verrucomicrobia show a similar picture, with only 10 defined species, i.e. Verrucomicrobium spinosum, four Prosthecobacter spp., Opitutus terrae, Rubritalea marinaand three Xiphinematobacter sp. (Ward-Rainey et al., 1995; Garrity et al., 2005). Although some representatives of the Acidobacteria and Verrucomicrobia have now been cultured from soil (Janssen et al., 2002; Sait et al., 2002; Stevenson et al., 2004; Davis et al., 2005), no isolates have as yet been obtained from the rhizosphere. Isolation directly from the rhizosphere is clearly needed to provide supportive data that these isolates are really rhizosphere-relevant organisms. Their availability would allow an assessment of their influences on plant growth, as well as of their interaction with plants and other soil microorganisms. Also, their involvement in important biogeochemical conversions, degradation of particular compounds and mobilization of nutrients would be facilitated. The members of the Acidobacteria and Verrucomicrobia that have previously been isolated from soil were slow growing, aerobic and heterotrophic bacteria (Janssen et al., 2002; Sait et al., 2002; Stevenson et al., 2004; Davis et al., 2005). Such characteristics are broadly distributed among rhizosphere isolates, which indicates that such isolates may thrive in the rhizosphere environment.
The abundance of the hitherto-unculturable bacteria recognized solely by their phylogeny in the rhizosphere (Kaiser et al., 2001; Kuske et al., 2002; Gremion et al., 2003; Schmalenberger & Tebbe, 2003; Graff & Conrad, 2005; Sharma et al., 2005; Stafford et al., 2005; Sanguin et al., 2006; Wang et al., 2007; Zhang et al., 2007; van Elsas et al., 2008) urges us to address their functional roles. However, information about their putative roles or interactions with the plant is as yet very sparse. Each of the organisms involved might, for instance, occupy a particular niche in the rhizosphere. It is a challenge to develop strategies – based on cultivation-dependent and -independent studies – that allow the exploration of the putative roles and function of the hitherto-unculturable microbiota in the rhizosphere, in particular, analysing their modes of interactions with plants and/or rhizosphere microorganisms.
Jump to…
Ecology of the hitherto-unculturable bacteria in the rhizosphere
If particular niches exist for the hitherto-unculturable bacteria in the rhizosphere, it is logical to assume that selective forces present within such niches act on these rhizosphere populations. Such a selection might be nutritional, dependent on the availability of oxygen or other electron acceptors, or otherwise. To our knowledge, no conclusive data have extensively shown the factors that influence the fate and behaviour of these populations in the rhizosphere. Therefore, the following points should be addressed to elucidate the roles of these organisms in the rhizosphere: (1) their general occurrence and numerical dominance, (2) the ecological conditions driving these populations, (3) in situ (local) metabolic activities and (4) their functional roles in the soil–plant ecosystem.
When comparing bulk and rhizosphere soil, the general occurrence and numerical abundances of the Acidobacteria, Verrucomicrobia and Planctomycetes are often variable, as demonstrated in culture-independent studies based on 16S rRNA gene detection (Dunbar et al., 1999; Kuske et al., 2002; Gremion et al., 2003; Filion et al., 2004; Stafford et al., 2005; Sanguin et al., 2006; De Cárcer et al., 2007; Singh et al., 2007; Zul et al., 2007; Kielak et al., 2008). Specifically, a higher prevalence of 16S rRNA gene clones affiliated with particular groups of Acidobacteria, especially those of groups 1, 2 and 3, was found in rhizospheres of lodgepole pine (Chow et al., 2002),Proteacea sp. (Stafford et al., 2005) and grass (Chow et al., 2002; Stafford et al., 2005; Singh et al., 2007) as compared with corresponding bulk soils. Also, a higher number of 16S rRNA gene clones of Verrucomicrobia was observed in rhizospheres of lodgepole pine, particularly subdivisions 2, 3 and 4 (Chow et al., 2002), Thlaspi caerulescens, subdivision 2 (Gremion et al., 2003) and Proteacea sp., subdivision 3 (Stafford et al., 2005) in comparison with the corresponding bulk soils. Besides, Planctomycetesrepresentatives were found in higher numbers in the rhizospheres of T. caerulescens than in the bulk soil, with 16S rRNA gene clones affiliating only with the Nostocoida limilula III cluster (Gremion et al., 2003) and Proteacea sp. (Stafford et al., 2005) (Table 1). In contrast, a study performed with different plants demonstrated higher numbers of Acidobacteria in a 16S rRNA gene clone library made from bulk soil DNA than in the ones made from rhizosphere DNA, although no information was given about the dominant groups in both soil compartments (Kielak et al., 2008). Also, the rhizosphere of maize plants showed lower numbers of Acidobacteria, Verrucomicrobia andPlanctomycetes than the corresponding bulk soil, and again no information was given about the dominant subdivisions (Sanguin et al., 2006). In T. caerulescens (Gremionet al., 2003), different Acidobacteria groups were found in bulk and rhizosphere soil, although the number of detected 16S rRNA genes was the same in both soil compartments. In this study, more clones were found to be affiliated with Acidobacteria subdivision 6 in the rhizosphere, whereas in the bulk soil more clones were found to be affiliated with Acidobacteria subdivision 1. Furthermore, the diversities of the Acidobacteria and Verrucomicrobia in the rhizosphere of Lolium perenne L. were large. Such diversities even approximated those of the total Proteobacteria (Singh et al., 2007).
Table 1. Abundance of Acidobacteria, Verrucomicrobia and Planctomycetes and presumed factors influencing community sizes in the rhizosphere
Plant species Soil type Approach Major observations made References
Higher abundance of the hitherto- uncultured groups found at Presumed factors influencing the Acidobacteria,Verrucomicrobia andPlanctomycetes communities
Bulk soil Rhizosphere
- Applied approaches for studying the bacterial community composition in described studies:
- A, 16S rRNA gene clone library; B, terminal restriction length polymorphism; C, amplified ribosomal DNA restriction analysis; D, 16S rRNA gene-based taxonomic microarray; E, PCR–thermal gradient gel electrophoresis; F, PCR–denaturating gradient gel electrophoresis; G, quantitative PCR; PCB, polychlorinated biphenyls; ND, not determined.
Bunchgrasses (Stipa hymenoides andHilaria jamesii); grass (Bromus tectorum) Calcareous loamy sand B ND ND • The composition ofAcidobacterium division members differed with soil depth and with the presence of plants or cyanobacterial crust on the top soil layer Kuske et al.(2002)
Thlaspi caerulescens Sandy loam A VerrucomicrobiaandPlanctomycetes • Plant roots exerted a strong selective pressure on the diversity of Acidobacteria, Verrucomicrobiaand Planctomycetesrepresentatives Gremion et al. (2003)
Conifer (Picea mariana) Not described C ND ND • Representatives of the division ofAcidobacteria were higher in number and diversity in the rhizosphere of healthy plants than of plants with black spruce •Verrucomicrobia representatives were only found in the rhizosphere of diseased black spruce plants Filion et al.(2004)
Proteacea species (Leucospermun truncatulum andLecadendron xanthoconus) Acid sand-stone-derived soil A Acidobacteria, VerrucomicrobiaandPlanctomycetes • Endemic communities in the rhizosphere differed per geographic location and soil covering vegetation type Stafford et al. (2005)
Maize (Zea mays cv. PR38a24) Sand–loam soil D Acidobacteria, VerrucomicrobiaandPlanctomycetes • Plant roots exerted a strong selective pressure on the diversity of Acidobacteria, Verrucomicrobiaand Planctomycetesrepresentatives Sanguin et al. (2006)
Willow (Salix vimanalis×schewerinii) Not described E ND ND • PCB had no effect on theAcidobacteria diversity in the rhizosphere of Willow De Cárcer et al. (2007)
Grass (Lolium perenne L.) Sand–silt soil A Acidobacteria • Acidobacterial communities were favoured in rhizosphere soil, but the major determinant of community composition was soil type Singh et al.(2007)
Different plant communities Endostagnic cambisol soil F Verrucomicrobia, PlanctomycetesandAcidobacteria • Plant species strongly influenced the bacterial community composition; season was identified as a second important factor. No effects caused by plant diversity or water content were observed Zul et al.(2007)
Different plant diversity treatments Loamy sand soil A Acidobacteria Verrucomicrobia • No detectable changes in acidobacterial or verrucomicrobial population diversity and structure were observed in response to different vegetation coverages Kielak et al.(2008)
Yew (Taxus×mediaand Taxus mairei) Not described A ND ND • Geographic origin is more important in shaping the bacterial community composition than plant species Hao et al.(2008)
Wild oat root (Avena fatua) Medium-texture loam G Acidobacteria, VerrucomicrobiaandPlanctomycetes • Rhizosphere effect increased numbers of Acidobacteria, Verrucomicrobia andPlanctomycetes member in rhizosphere soil when compared with the respective bulk soil • The number of Acidobacteria, Verrucomicrobia andPlancomycetes also differed among root tip, root hairs and mature root DeAngelis et al. (2009)
The numerical abundance of members of the Acidobacteria, Verrucomicrobia and Planctomycetes is not only dependent on differences between bulk and rhizosphere soil but also changes are incited upon root development. In a microcosm experiment performed with wild oat roots, members of the Acidobacteria, Verrucomicrobia andPlanctomycetes occurred at a higher number, as demonstrated by the higher number of 16S rRNA gene copies in the rhizosphere of adult plants than in the bulk soil (DeAngelis et al., 2009). Besides, there was a greater abundance of Acidobacteria 16S rRNA genes at the surface of mature roots than at root hairs and tips; thePlanctomycetes showed higher 16S rRNA gene abundance at the surface of mature roots and root tips, than at the root hairs, and the Verrucomicrobia, although having a lower abundance at the root tips, had greater abundance at the surface of mature roots and root hairs than in the corresponding bulk soil (DeAngelis et al., 2009).
Clearly, the high diversity of acidobacterial and verrucomicrobial species indicates that different bacterial populations are favoured by different selective forces exerted in the rhizosphere. At the microhabitat level, such forces can be different across space and time, thus allowing the emergence of diverse populations across each phylum. An important question here is which selective forces influence the acidobacterial, verrucomicrobial and Planctomycetales communities in the rhizosphere. On the basis of incidental data, this question has been difficult to answer. Thus, no consensus was found about which ecological factors would select for these organisms in the rhizosphere. Recent studies demonstrated that different factors select distinct members of these groups (Table 1). For instance, plant species (Chow et al., 2002; Gremionet al., 2003; Sanguin et al., 2006), soil type (Singh et al., 2007), field history (Kielak et al., 2008) and different abiotic factors (Stafford et al., 2005; Hao et al., 2008) have all been implicated as selective factors that differentially affect these groups (Table 1).
Moreover, another key question is whether we can actually show to what extent hitherto-uncultured organisms, for example members of the Acidobacteria, are part of the metabolically active community (derived from the higher abundances in RNA vs. DNA extracts) in the rhizosphere and how this is temporally organized. In the metal-hyperaccumulating plant T. caerulescens, a major discrepancy in abundance was found between the metabolically active and the total Acidobacteria (Gremion et al., 2003). In this study, it was indicated that only a subset of the Acidobacteria might be active in the rhizosphere, whereas the majority may be inactive or dormant. On the other hand, in a study using a culture-independent approach to search for 16S rRNA gene sequences in DNA and RNA extracts from the chestnut tree (Castanea crenata) rhizosphere (Lee et al., 2008), Acidobacteria were found to be not only numerically dominant but also metabolically active, implying that these Acidobacteria may play a key role in this habitat. Additionally, in a metagenomic study conducted on the rhizosphere of Erica andevalensis adapted to AMD, it was demonstrated that in this acid environment,Acidobacteria of subdivision 1 were metabolically active (Mirete et al., 2007). The authors also found that four of 13 clones with nickel resistance genes most likely were from representatives of Acidobacteria subdivision 1 (Mirete et al., 2007). However, it is unclear whether these genes were actually expressed in the environment and whether they play key roles in the heavy metal resistance of E. andevalensis.
Jump to…
Methods to improve bacterial and archaeal culturability
Recently, several successes in the culturing of the hitherto-uncultured bacteria were reported (Table 2). Key factors that presumably influence bacterial colony formation on solid media were found to be the type of growth medium, the incubation conditions (temperature, CO2 concentration and time of incubation) and several factors that are known to reduce oxidative stress (e.g. the presence of catalase or pyruvate) (Stevenson et al., 2004; Van Overbeek et al., 2004; Sangwan et al., 2005; Eichorst et al., 2007). It is a well-known fact that medium composition will determine which fraction of the microbial community is recoverable from the environment. Often, the medium itself limits bacterial growth due to the presence of excessive amounts of nutrients that do not reflect environmental conditions (Janssen et al., 2002; Sait et al., 2002). This phenomenon, known as ‘substrate-accelerated death’, has first been observed when organisms growing under oligotrophic conditions are transferred to high substrate concentrations (Postgate & Hunter, 1964; Straskrabová, 1983). It is a key issue, given the roughly low substrate concentrations in the rhizosphere compartment. Therefore, to overcome this limitation, novel media are designed to match conditions in the environment in which some of the key facets (e.g. amount of nutrients, nutrient composition, the presence of trace elements and pH) are adjusted for optimized bacterial growth. For instance, the use of soil-extract agar medium (Pochon & Tardieu, 1962; Hamaki et al., 2005) or of dilute standard media, both aiming to reduce nutrient levels in order to avoid substrate-accelerated death, has allowed the isolation of bacterial groups that had been regarded as uncultured only a short while ago (Joseph et al., 2003; Davis et al., 2005). Also, the presence of fast-growing bacteria that inhibit colony formation of slower growing ones, especially on nutrient-rich media, affects the recovery of hard-to-culture organisms. Media with low nutrient levels, in combination with long incubation periods at relatively low temperatures, will allow colony formation by bacteria that have low growth rates. The approach has been successful in revealing a range of novel organisms in various recent studies (Janssen et al., 2002; Rappe et al., 2002; Sait et al., 2002; Davis et al., 2005; Miteva & Brenchley, 2005; Sangwan et al., 2005; Eichorst et al., 2007). Hence, this allowed the cultivation from soil of a range of novel organisms such as bacteria belonging to the following groups: Chloroflexi, Planctomycetes, Acidobacteria, Verrucomicrobia and some Archaea representatives (Table 2). However, such approaches have seldom been used with rhizosphere samples, and only one reference was found reporting cultivation of hard-to-culture microorganisms from rhizosphere samples (Simon et al., 2005).
Table 2. Factors hampering bacterial growth upon isolation from natural environments and proposed approaches to improve culturability
Factors that limit growth of the hitherto-uncultured bacteria Proposed approaches to improve culturability Phylogeny of novel isolates that were obtained Environments from where novel isolates were obtained References
Inability to grow at high nutrient concentrations and overgrowth by faster growing species • Reduce nutrient availability in growth medium • Apply a longer incubation time Actinobacteria, Acidobacteria, Proteobacteria, Verrucomicrobia, Gammaproteobacteria, SAR11 clade,Gemmatimonadetes, Chloroflexi, Planctomycetes, CFB group • Bulk soil Janssen et al. (2002);Sait et al. (2002);Davis et al. (2005);Sangwan et al. (2005);Eichorst et al. (2007)
• Seawater Rappe et al. (2002);Miteva & Brenchley (2005)
Media selectiveness for particular groups of microorganisms • Development of media that select for a different or a broader spectrum of microorganisms Acidobacteria, Gemmatimonadetes, Chloroflexi, Alphaproteobacteria, Betaproteobacteria, Planctomycetes, Gammaproteobacteria, Actinobacteria, • Bulk soil Joseph et al. (2003);Davis et al. (2005);Hamaki et al. (2005)
Compounds inhibiting bacterial growth • Diffusion or dilution of growth-inhibiting compounds, for example, by making use of diffusion chambers Deltaproteobacteria, Verrucomicrobia, Spirochaetes, Acidobacteria, Planctomycetes, Proteobacteria, Bacteroidetes • Marine sediment Bollmann et al. (2007)
• Fresh water sediment Tamaki et al. (2005)
• Application of alternative solidifying agents, for example gellan gun instead of agar
Syntrophic growth or requirement for growth factors produced by other microorganisms • Incubation of environmental samples in diffusion chambers • Addition of syntrophic bacteria in enrichment cultures Deltaproteobacteria, Verrucomicrobia, Spirochaetes, Acidobacteria Rice Cluster-I methanogen group Archaea • Rice paddy field Sakai et al. (2007)
• Fresh water sediment Bollmann et al. (2007)
• Bulk soil Cavaletti et al. (2006)
• Rhizosphere Simon et al. (2005)
CO2 tension during incubation • Increase CO2 tension during incubation Verrucomicrobia, Acidobacteria • Bulk soil Stevenson et al.(2004); Sangwan et al.(2005); Eichorst et al.(2007)
Low abundance in environmental samples • Single-cell detection combined with parallel microbial cultivation • High-throughput dilution to extinction cultivation method • Micro-Petri dish • Bulk soil Zengler et al. (2005)
• Seawater Rappe et al. (2002)
• Fresh water Ingham et al. (2007)
Formation of colonies that are undetectable by the unarmed eye • Colony identification by FISH in microtitre plates • Colony identification by hybridization on nylon membranes • Microcolony cultivation Verrucomicrobia, Candidate division TM7, Acidobacteria • Bulk soil Stevenson et al.(2004); Ferrari et al.(2005); Sangwan et al.(2005)
• Sea water Rappe et al. (2002)
Restriction of bacterial growth upon the presence of reactive oxygen species • Reduction of oxidative stress by addition of oxidation-protective agents Acidobacteria • Bulk soil Stevenson et al.(2004)
Lack of ability to grow under specific circumstances, i.e. growth on solid or in a liquid substrate • Screening for isolates in both liquid and solid media Acidobacteria, Verrucomicrobia, Gemmatimonadetes, Proteobacteriaand CFB group • Sea food, salt-processed food, raw vegetables Shigematsu et al.(2007)
• Seawater Miteva & Brenchley (2005); Simu et al.(2005)
• Bulk soil Schoenborn et al.(2004)
Besides, the inadvertent presence of growth-inhibitory compounds in agar media may prevent the formation of colonies by particular bacteria. The use of gellan gum as an alternative solidifying agent was shown to allow the cultivation of many hitherto-unculturable bacteria (Tamaki et al., 2005), as the agar might actually be inhibitory to some groups of microorganisms, for example, those from freshwater. Also, incubation of samples (in this case from marine sediments) in diffusion chambers, which allows dilution of inhibitory compounds produced by bacteria during growth, permitted the growth of novel bacterial groups (Bollmann et al., 2007) (Table 2). Another reason why many bacteria cannot be cultured is the obligate growth of bacteria in consortia (Sakai et al., 2007). Thus, the growth of individual species in these consortia relies on the growth of others, for example when bacteria feed on compounds produced by others (Simon et al., 2005) or require growth factors produced by other bacteria (Bollmann et al., 2007). Therefore, it will be relevant to screen for unknown species in clone libraries made from bacterial mixtures, for example, by plating undiluted rhizosphere samples.
Jump to…
Towards the as-yet-uncultured microbial populations in the rhizosphere and their basic ecology
The recovery of the broadest spectrum of species diversity from the rhizosphere will offer a new insight into hitherto unculturable populations and their functions, as demonstrated with the recovery of novel nickel resistance genes from heather (E. andevalensis) rhizosphere metagenome (Mirete et al., 2007). A major focus should be placed on those bacterial phyla that are presumably abundant in the rhizosphere and that have so far only been described via culture-independent approaches, i.e. theAcidobacteria, Verrucomicrobia and Planctomycetes. Several studies have indicated strategies to improve culturability in natural ecosystems (Table 2), but only one has so far been addressed in the rhizosphere, in this case by improving the culturability of Archaea species (Simon et al., 2005).
Improving the cultivation of Acidobacteria and Verrucomicrobia from the rhizosphere
The rhizosphere clearly is a selective habitat for different groups of microorganisms in soil and this selectivity comes about as a result of a plethora of beneficial and stress conditions imposed on its inhabitants. Analysis of the factors that shape rhizosphere conditions might help to design methods for the recovery of a broad range of novel isolates. The following factors should be considered as relevant for the design of such methods: (1) the provision of the (solid) substrate for bacterial growth; (2) the nutritional status of the rhizosphere, which is often oligotrophic, although pulse-wise receiving nutrients; (3) the high partial CO2 and low partial O2 pressures (due to bacterial and root respiration); (4) the temporary and fluctuating lowering of the pH and (5) the presence of signalling and other (eventually toxic) molecules of root and bacterial origin. Given the fact that no single medium or technique is likely to solve the culturability issue all at once, here, a polyphasic approach is proposed, which should apply parallel cultivation systems in an attempt to yield as many hitherto-uncultured bacteria from the rhizosphere as possible. Key to such a polyphasic approach will be:
- 1
Use of low-nutrient media in combination with long incubation periods to avoid substrate-induced death and overgrowth of fast growers, thus supporting the growth of slower ones. - 2
Inclusion of incubation at elevated carbon dioxide tension levels. - 3
Amendment with agents in the growth medium that protect cells from reactive oxygen species produced during their metabolism or present as a result of autoclaving. - 4
Amendment of media with rhizosphere extract to mimic the rhizosphere environment, with an emphasis on the presence of available nutrients and signalling molecules for growth. - 5
Parallel use of solid and liquid media – the rhizosphere offers both a solid substrate (root surface and soil matrix) and an aqueous phase (water film surrounding soil particles and water-filled soil pores) for bacterial growth; thus, the use of a combination of solid and liquid media will support the outgrowth of microorganisms with sessile and benthic lifestyles.
Jump to…
Conclusions
Rhizosphere communities are important for key plant processes such as nutrient acquisition, protection against soil-borne diseases and plant development, as the rhizosphere is the transitional zone in between bulk soil and plant roots. The key functions of rhizosphere bacterial communities, for instance nitrogen fixation (Canbolat et al., 2006), phosphate solubilization (Kohler et al., 2006) and plant health protection (Garbeva et al., 2004a), have been described, mainly for well-characterized culturable species.
In this review, we show that among the groups of bacteria highest in density in the rhizosphere environment, the Acidobacteria, Verrucomicrobia and Planctomycetes are the ones whose functions remain to be unveiled. So far, these three groups have not been cultivated from this environment. Therefore, most of the information that emerged on these taxa in the rhizosphere was from measurements on the 16S rRNA gene level. The only exception is a metagenomics study conducted in the rhizosphere of plants adapted to AMD (Mirete et al., 2007). However, in this study, not all modes of action of the novel genes found could be elucidated nor was it shown that these genes were actually active in the rhizosphere environment.
One way to circumvent the hurdles of culture-independent techniques (Van Elsas et al., 2008) is the development of approaches that improve culturability, here, with a special focus on the rhizosphere environment. Several studies already aimed at the isolation of novel species from bulk soil, with variable successes in the recovery ofAcidobacteria and Verrucomicrobia. Although these two groups have shown their recalcitrance to cultivation from the rhizosphere environment, it is likely to presume that they can be isolated using approaches directed to recover the widest possible range of bacteria. We propose the use of a polyphasic approach comprising low-nutrient solid and liquid media with the inclusion of oxidative stress-protective agents and/or rhizosphere extract, and long incubation periods at elevated carbon dioxide concentrations. The exploration of hitherto-uncultured organisms – following their growth to purity – will clearly simplify an assessment of their interactions with other organisms, which cannot be achieved via culture-independent approaches.
Moreover, there is a lack of studies that merge culture-dependent with advanced culture-independent studies. Such a combination of approaches will greatly increase our analytical power in order to answer one of the ultimate questions in microbial ecology –‘Which are the ecological niches, putative roles, functions and modes of interactions of the hitherto-uncultured microbiota?’
http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6941.2009.00702.x/full
References
- Andreote FD, Mendes R, Dini-Andreote F et al. (2008) Transgenic tobacco revealing altered bacterial diversity in the rhizosphere during early plant development. Antonie van Leeuwenhoek 93: 415–424.
- Belimov AA, Dodd IC, Hontzeas N, Theobald JC, Safronova VI & Davies WJ (2009) Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol 181:413–423.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(284K)
- References
- Web of Science® Times Cited: 4
- Benedict MA & Frelich LE (2008) Site factors affecting black ash ring growth in northern Minnesota. Forest Ecol Manag 255: 3489–3493.
- CrossRef,
- Web of Science® Times Cited: 1
- Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A & Hallmann J (2005) Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol 51: 215–229.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(393K)
- References
- Web of Science® Times Cited: 55
- Bharathkumar S, RameshKumar N, Paul D, Prabavathy VR & Nair S (2008) Characterization of the predominant bacterial population of different mangrove rhizosphere soils using 16S rRNA gene-based single-strand conformation polymorphism (SSCP). World J Microb Biot 24: 387–394.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 2
- Biryukova OV, Fedorak PM & Quideau SA (2007) Biodegradation of naphthenic acids by rhizosphere microorganisms. Chemosphere 67:2058–2064.
- Bollmann A, Lewis K & Epstein SS (2007) Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microb 73: 6386–6390.
- Bryant DA, Garcia Costas AM, Maresca JA et al. (2007) Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317: 523–526.
- Canbolat MY, Bilen S, Cakmakci R, Sahin F & Aydin A (2006) Effect of plant growth-promoting bacteria and soil compaction on barley seedling growth, nutrient uptake, soil properties and rhizosphere microflora. Biol Fert Soils 42: 350–357.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 12
- Cavaletti L, Monciardini P, Bamonte R, Schumann P, Ronde M, Sosio M & Donadio S (2006) New lineage of filamentous, spore-forming, gram-positive bacteria from soil. Appl Environ Microb 72: 4360–4369.
- Chow ML, Radomski CC, McDermott JM, Davies J & Axelrood PE (2002) Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol Ecol 42: 347–357.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(342K)
- References
- Web of Science® Times Cited: 37
- Cole JR, Chai B, Farris RJ et al. (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172.
- Colmer TD (2003) Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ 26: 17–36.
- CAS,
- Web of Science® Times Cited: 136
- Costa R, Gomes NCM, Krogerrecklenfort E, Opelt K, Berg G & Smalla K (2007) Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ Microbiol 9:2260–2273.
- Davis KER, Joseph SJ & Janssen PH (2005) Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microb 71: 826–834.
- DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE & Firestone MK (2009) Selective progressive response of soil microbial community to wild oat roots. ISME J 3: 168–178.
- De Cárcer DA, Martin M, MacKova M, MacEk T, Karlson U & Rivilla R (2007) The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J 1: 215–223.
- DeLong EF (2005) Microbial community genomics in the ocean. Nat Rev Microbiol 3: 459–469.
- DeLong EF, Preston CM, Mincer T et al. (2006) Community genomics among stratified microbial assemblages in the ocean's inferior.Science 311: 496–503.
- Dunbar J, Takala S, Barns SM, Davis JA & Kuske CR (1999) Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microb 65: 1662–1669.
- Eichorst SA, Breznak JA & Schmidt TM (2007) Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl Environ Microb 73: 2708–2717.
- Elliott ML, McInroy JA, Xiong K, Kim JH, Skipper HD & Guertal EA (2008) Taxonomic diversity of rhizosphere bacteria in golf course putting greens at representative sites in the Southeastern United States. HortScience 43: 514–518.
- Web of Science® Times Cited: 1
- Ferrari BC, Binnerup SJ & Gillings M (2005) Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microb 71: 8714–8720.
- Fierer N, Bradford MA & Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 59
- Filion M, Hamelin RC, Bernier L & St-Arnaud M (2004) Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microb 70: 3541–3551.
- Flores-Vargas RD & O'Hara GW (2006) Isolation and characterization of rhizosphere bacteria with potential for biological control of weeds in vineyards. J Appl Microbiol 100: 946–954.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(263K)
- References
- Web of Science® Times Cited: 10
- Földes T, Bánhegyi I, Herpai Z, Varga L & Szigeti J (2000) Isolation of Bacillus strains from the rhizosphere of cereals and in vitro screening for antagonism against phytopathogenic, food-borne pathogenic and spoilage micro-organisms. J Appl Microbiol 89: 840–846.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(119K)
- References
- Web of Science® Times Cited: 25
- Garbeva P, Van Veen JA & Van Elsas JD (2004a) Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, ofPseudomonas species in soil from different agricultural regimes. FEMS Microbiol Ecol 47: 51–64.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(429K)
- References
- Web of Science® Times Cited: 43
- Garbeva P, Van Veen JA & Van Elsas JD (2004b) Microbial diversity in soils: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42: 243–270.
- Garbeva P, Van Elsas JD & Van Veen JA (2008) Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 302: 19–32.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 5
- Garrity GM, Bell JA & Lilburn TG (2005) Taxonomic outline of the prokaryotes. Bergey's Manual of Systematic Bacteriology, Second Edition. Release 5.0 May 2004 (GarrityGM, editor-in-chief), pp. 1–399. Springer-Verlag, NY.
- Gaumont-Guay D, Black TA, Barr AG, Jassal RS & Nesic Z (2008) Biophysical controls on rhizospheric and heterotrophic components of soil respiration in a boreal black spruce stand. Tree Physiol 28: 161–171.
- Graff A & Conrad R (2005) Impact of flooding on soil bacterial communities associated with poplar (Populus sp.) trees. FEMS Microbiol Ecol 53: 401–415.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(362K)
- References
- Web of Science® Times Cited: 8
- Gremion F, Chatzinotas A & Harms H (2003) Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ Microbiol 5:896–907.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(337K)
- References
- Web of Science® Times Cited: 36
- Haggag WM (2007) Colonization of exopolysaccharide-producing Paenibacillus polymyxa on peanut roots for enhancing resistance against crown rot disease. Afr J Biotechnol 6: 1568–1577.
- CAS,
- Web of Science® Times Cited: 3
- Hamaki T, Suzuki M, Fudou R et al. (2005) Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J Biosci Bioeng 99:485–492.
- Hameeda B, Harini G, Rupela OP, Wani SP & Reddy G (2008) Growth promotion of maize by phosphate-solubilizing bacteria isolated from composts and macrofauna. Microbiol Res 163: 234–242.
- Hao DC, Ge GB & Yang L (2008) Bacterial diversity of Taxus rhizosphere: culture-independent and culture-dependent approaches. FEMS Microbiol Let 284: 204–212.
- Hardoim PR, Van Overbeek LS & Elsas JDv (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16: 463–471.
- Herrmann M, Saunders AM & Schramm A (2008) Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microb 74: 3279–3283.
- Hiltner L (1904) Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Gründüngung und Brache. Arbeiten der Deutschen Landwirtschaftlichen Gesellschaft 98: 59–78.
- Hinsinger P (1998) How do plant roots acquire mineral nutrients? Chemical processes involved in the rhizosphere. Adv Agron 64: 225–265.
- Hinsinger P, Gobran GR, Gregory PJ & Wenzel WW (2005) Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytologist 168: 293–303.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(391K)
- References
- Web of Science® Times Cited: 67
- Huisman OC (1982) Interrelations of root growth dynamics to epidemiology of root-invading fungi. Annu Rev Phytopathol 20: 303–327.
- CrossRef,
- Web of Science® Times Cited: 67
- Ingham CJ, Sprenkels A, Bomer J, Molenaar D, Van Den Berg A, Van Hylckama Vlieg JET & De Vos WM (2007) The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. P Natl Acad Sci USA 104: 18217–18222.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 23,
- ADS
- Jackson RB, Canadell J, Ehleringer JR, Mooney HA, Sala OE & Schulze ED (1996) A global analysis of root distributions for terrestrial biomes. Oecologia 108: 389–411.
- CrossRef,
- Web of Science® Times Cited: 502
- Janssen PH, Yates PS, Grinton BE, Taylor PM & Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microb 68: 2391–2396.
- Jiménez-Esquilín AE & Roane TM (2005) Antifungal activities of actinomycete strains associated with high-altitude sagebrush rhizosphere. J Ind Microbiol Biot 32: 378–381.
- Jobbágy EG & Jackson RB (2000) The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol Appl 10:423–436.
- 2.0.CO;2']CrossRef,
- Web of Science® Times Cited: 369
- Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA & Janssen PH (2003) Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microb 69: 7210–7215.
- Jossi M, Fromin N, Tarnawski S, Kohler F, Gillet F, Aragno M & Hamelin J (2006) How elevated pCO2 modifies total and metabolically active bacterial communities in the rhizosphere of two perennial grasses grown under field conditions. FEMS Microbiol Ecol 55: 339–350.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(341K)
- References
- Web of Science® Times Cited: 10
- Kaci Y, Heyraud A, Barakat M & Heulin T (2005) Isolation and identification of an EPS-producing Rhizobium strain from arid soil (Algeria):characterization of its EPS and the effect of inoculation on wheat rhizosphere soil structure. Res Microbiol 156: 522–531.
- Kaiser O, Puhler A & Selbitschka W (2001) Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb Ecol 42: 136–149.
- Kielak A, Pijl AS, Van Veen JA & Kowalchuk GA (2008) Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol Ecol 63: 372–382.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(146K)
- References
- Web of Science® Times Cited: 8
- Kiely PD, Haynes JM, Higgins CH, Franks A, Mark GL, Morrissey JP & O'Gara F (2006) Exploiting new systems-based strategies to elucidate plant–bacterial interactions in the rhizosphere. Microb Ecol 51: 257–266.
- Koch IH, Gich F, Dunfield PF & Overmann J (2008) Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int J Syst Evol Micr 58: 1114–1122.
- Kohler J, Caravaca F, Carrasco L & Roldan A (2006) Contribution of Pseudomonas mendocina and Glomus intraradices to aggregate stabilization and promotion of biological fertility in rhizosphere soil of lettuce plants under field conditions. Soil Use Manage 22: 298–304.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(505K)
- References
- Web of Science® Times Cited: 11
- Kotani-Tanoi T, Nishiyama M, Otsuka S & Senoo K (2007) Single particle analysis reveals that bacterial community structures are semi-specific to the type of soil particle. Soil Sci Plant Nutr 53: 740–743.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(94K)
- References
- Web of Science® Times Cited: 2
- Krivtsov V, Garside A, Brendler A, Liddell K, Griffiths BS & Staines HJ (2007) A study of population numbers and ecological interactions of soil and forest floor microfauna. Anim Biol 57: 467–484.
- CrossRef,
- Web of Science® Times Cited: 2
- Kuske CR, Ticknor LO, Miller ME, Dunbar JM, Davis JA, Barns SM & Belnap J (2002) Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl Environ Microb 68: 1854–1863.
- Lee KH & Song HG (2007) Growth promotion of tomato seedlings by application of Bacillus sp. isolated from rhizosphere. Korean J Microbiol 43: 279–284.
- CrossRef
- Lee S-H, Ka J-O & Cho J-C (2008) Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol Let 285: 263–269.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(172K)
- References
- Web of Science® Times Cited: 5
- Lipiec J, Walczak R, Witkowska-Walczak B, Nosalewicz A, Sowinska-Jurkiewicz A & Sawinski C (2007) The effect of aggregate size on water retention and pore structure of two silt loam soils of different genesis. Soil Till Res 97: 239–246.
- CrossRef,
- Web of Science® Times Cited: 2
- Ludwig W, Strunk O, Westram R et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
- Mirete S, De Figueras CG & Gonzalez-Pastor JE (2007) Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl Environ Microbiol 73: 6001–6011.
- Miteva VI & Brenchley JE (2005) Detection and isolation of ultrasmall microorganisms from a 120 000-year-old Greenland glacier ice core.Appl Environ Microb 71: 7806–7818.
- Narasimhan K, Basheer C, Bajic VB & Swarup S (2003) Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132: 146–153.
- Nelson DR & Mele PM (2006) The impact of crop residue amendments and lime on microbial community structure and nitrogen-fixing bacteria in the wheat rhizosphere. Aust J Soil Res 44: 319–329.
- CrossRef,
- Web of Science® Times Cited: 8
- Nichols D (2007) Cultivation gives context to the microbial ecologist. FEMS Microbiol Ecol 60: 351–357.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(87K)
- References
- Web of Science® Times Cited: 13
- Nicolaisen MH, Risgaard-Petersen N, Revsbech NP, Reichardt W & Ramsing NB (2004) Nitrification–denitrification dynamics and community structure of ammonia oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol Ecol 49: 359–369.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(388K)
- References
- Web of Science® Times Cited: 20
- Pandey A, Palni LMS & Bisht D (2001) Dominant fungi in the rhizosphere of established tea bushes and their interaction with the dominant bacteria under in situ conditions. Microbiol Res 156: 377–382.
- Passioura JB (1991) Soil structure and plant growth. Aust J Soil Res 29: 717–728.
- CrossRef,
- Web of Science® Times Cited: 89
- Phillips DA, Ferris H, Cook DR & Strong DR (2003) Molecular control points in rhizosphere food webs. Ecology 84: 816–826.
- 2.0.CO;2']CrossRef,
- Web of Science® Times Cited: 26
- Pochon J & Tardieu P (1962) Techniques d'Analyse en Microbiologie du Sol. Editions de la Tourella, Paris.
- Poonguzhali S, Madhaiyan M & Sa T (2006) Cultivation-dependent characterization of rhizobacterial communities from field grown Chinese cabbage Brassica campestris ssp pekinensis and screening of traits for potential plant growth promotion. Plant Soil 286: 167–180.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 7
- Postgate JR & Hunter JR (1964) Accelerated death of Aerobacter aerogenes starved in the presence of growth limiting substrates. J Gen Microbiol 34: 459–473.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J & Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 21: 7188–7196.
- Rappe MS, Connon SA, Vergin KL & Giovannoni SJ (2002) Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633.
- Romanenko LA, Uchino M, Tanaka N, Frolova GM, Slinkina NN & Mikhailov VV (2007) Occurrence and antagonistic potential ofStenotrophomonas strains isolated from deep-sea invertebrates. Arch Microbiol 189: 1–8.
- PubMed,
- Web of Science® Times Cited: 6
- Sait M, Hugenholtz P & Janssen PH (2002) Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4: 654–666.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(757K)
- References
- Web of Science® Times Cited: 122
- Sakai S, Imachi H, Sekiguchi Y, Ohashi A, Harada H & Kamagata Y (2007) Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster rice cluster. Appl Environ Microb 73: 4326–4331.
- Sanguin H, Remenant B, Dechesne A et al. (2006) Potential of a 16S rRNA-based taxonomic microarray for analyzing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl Environ Microb 72: 4302–4312.
- Sangwan P, Kovac S, Davis KER, Sait M & Janssen PH (2005) Detection and cultivation of soil Verrucomicrobia. Appl Environ Microb 71:8402–8410.
- Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF & Hutchinson CR (2005) Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microb 71:4840–4849.
- Schmalenberger A & Tebbe CC (2003) Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol Ecol 12: 251–261.
- Schoenborn L, Yates PS, Grinton BE, Hugenholtz P & Janssen PH (2004) Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microb 70: 4363–4366.
- Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H & Kandeler E (2001) Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microb 67: 4215–4224.
- Sfalanga A, Di Cello F, Mugnai L, Tegli S, Fani R & Surico G (1999) Isolation and characterisation of a new antagonistic Burkholderia strain from the rhizosphere of healthy tomato plants. Res Microbiol 150: 45–59.
- Sharma S, Aneja MK, Mayer J, Munch JC & Schloter M (2005) Characterization of bacterial community structure in rhizosphere soil of grain legumes. Microb Ecol 49: 407–415.
- Shigematsu T, Ueno S, Tsuchida Y, Hayashi M, Okonogi H, Masaki H & Fujii T (2007) Comparative analyses of viable bacterial counts in foods and seawater under microplate based liquid- and conventional agar plate cultivation: increased culturability of marine bacteria under liquid cultivation. Biosci Biotech Bioch 71: 3093–3097.
- Simon HM, Jahn CE, Bergerud LT, Sliwinski MK, Weimer PJ, Willis DK & Goodman RM (2005) Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl Environ Microb 71: 4751–4760.
- Simu K, Holmfeldt K, Zweifel UL & Hagstrom A (2005) Culturability and coexistence of colony-forming and single-cell marine bacterioplankton. Appl Environ Microb 71: 4793–4800.
- Singh BK, Munro S, Potts JM & Millard P (2007) Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl Soil Ecol 36: 147–155.
- CrossRef,
- Web of Science® Times Cited: 15
- Stafford WHL, Baker GC, Brown SA, Burton SG & Cowan DA (2005) Bacterial diversity in the rhizosphere of Proteaceae species. Environ Microbiol 7: 1755–1768.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(415K)
- References
- Web of Science® Times Cited: 7
- Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM & Breznak JA (2004) New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microb 70: 4748–4755.
- Stewart JB, Moran CJ & Wood JT (1999) Macropore sheath: quantification of plant root and soil macropore association. Plant Soil 211:59–67.
- CAS,
- Web of Science® Times Cited: 34
- Tamaki H, Sekiguchi Y, Hanada S, Nakamura K, Nomura N, Matsumura M & Kamagata Y (2005) Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl Environ Microb 71:2162–2169.
- Tringe SG, Von Mering C, Kobayashi A et al. (2005) Comparative metagenomics of microbial communities. Science 308: 554–557.
- Turpault MP, Gobran GR & Bonnaud P (2007) Temporal variations of rhizosphere and bulk soil chemistry in a Douglas fir stand. Geoderma137: 490–496.
- Tyson GW, Chapman J, Hugenholtz P et al. (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43.
- Vaishampayan PA, Kanekar PP & Dhakephalkar PK (2007) Isolation and characterization of Arthrobacter sp. strain MCM B-436, an atrazine-degrading bacterium, from rhizospheric soil. Int Biodeter Biodegr 60: 273–278.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 2
- Vandenkoornhuyse P, Mahe S, Ineson P et al. (2007) Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. P Natl Acad Sci USA 104: 16970–16975.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 18,
- ADS
- Van Elsas JD, Speksnijder AJ & Van Overbeek LS (2008) A procedure for the metagenomics exploration of disease-suppressive soils. J Microbiol Meth 75: 515–522.
- Van Overbeek LS, Bergervoet JHW, Jacobs FHH & Van Elsas JD (2004) The low-temperature-induced viable-but-nonculturable state affects the virulence of Ralstonia solanacearum Biovar 2. Phytopathology 94: 463–469.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 12
- Venter JC, Remington K, Heidelberg JF et al. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 1047,
- ADS
- Villadas PJ, Fernandez-Lopez M, Ramirez-Saad H & Toro N (2007) Rhizosphere-bacterial community in Eperua falcata (Caesalpiniaceae) a putative nitrogen-fixing tree from French Guiana rainforest. Microb Ecol 53: 317–327.
- Wang M, Chen JK & Li B (2007) Characterization of bacterial community structure and diversity in rhizosphere soils of three plants in rapidly changing salt marshes using 16S rDNA. Pedosphere 17: 545–556.
- Ward-Rainey N, Rainey FA, Schiesner H & Stackebrandt E (1995) Assignment of hitherto unidentified 165 rDNA species to a main line of descent within the domain Bacteria. Microbiology 141: 3247–3250.
- Watt M, Hugenholtz P, White R & Vinall K (2006a) Numbers and locations of native bacteria on field-grown wheat roots quantified by fluorescence in situ hybridization (FISH). Environ Microbiol 8: 871–884.
Direct Link:- Abstract
- Full Article (HTML)
- PDF(863K)
- References
- Web of Science® Times Cited: 16
- Watt M, Silk WK & Passioura JB (2006b) Rates of root and organism growth, soil conditions, and temporal and spatial development of the rhizosphere. Ann Bot 97: 839–855.
- CrossRef,
- PubMed,
- Web of Science® Times Cited: 33
- Whalley WR, Watts CW, Gregory AS, Mooney SJ, Clark LJ & Whitmore AP (2008) The effect of soil strength on the yield of wheat. Plant Soil 306: 237–247.
- CrossRef,
- CAS,
- Web of Science® Times Cited: 5
- Zengler K, Walcher M, Clark G et al. (2005) High-throughput cultivation of microorganisms using microcapsules. Meth Enzymol 397:124–130.
- Zhang HB, Shi W, Yang MX, Sha T & Zhao ZW (2007) Bacterial diversity at different depths in lead-zinc mine tailings as revealed by 16S rRNA gene libraries. J Microbiol 45: 479–484.
- Zul D, Denzel S, Kotz A & Overmann J (2007) Effects of plant biomass, plant diversity, and water content on bacterial communities in soil lysimeters: implications for the determinants of bacterial diversity. Appl Environ Microb 73: 6916–6929.





